With most of the code ready for scoring target protein sequences for peptide binding to selected MHC alleles we can move to the web application part.
We will implement a simple layout very similar to those used for our previous examples and applications.
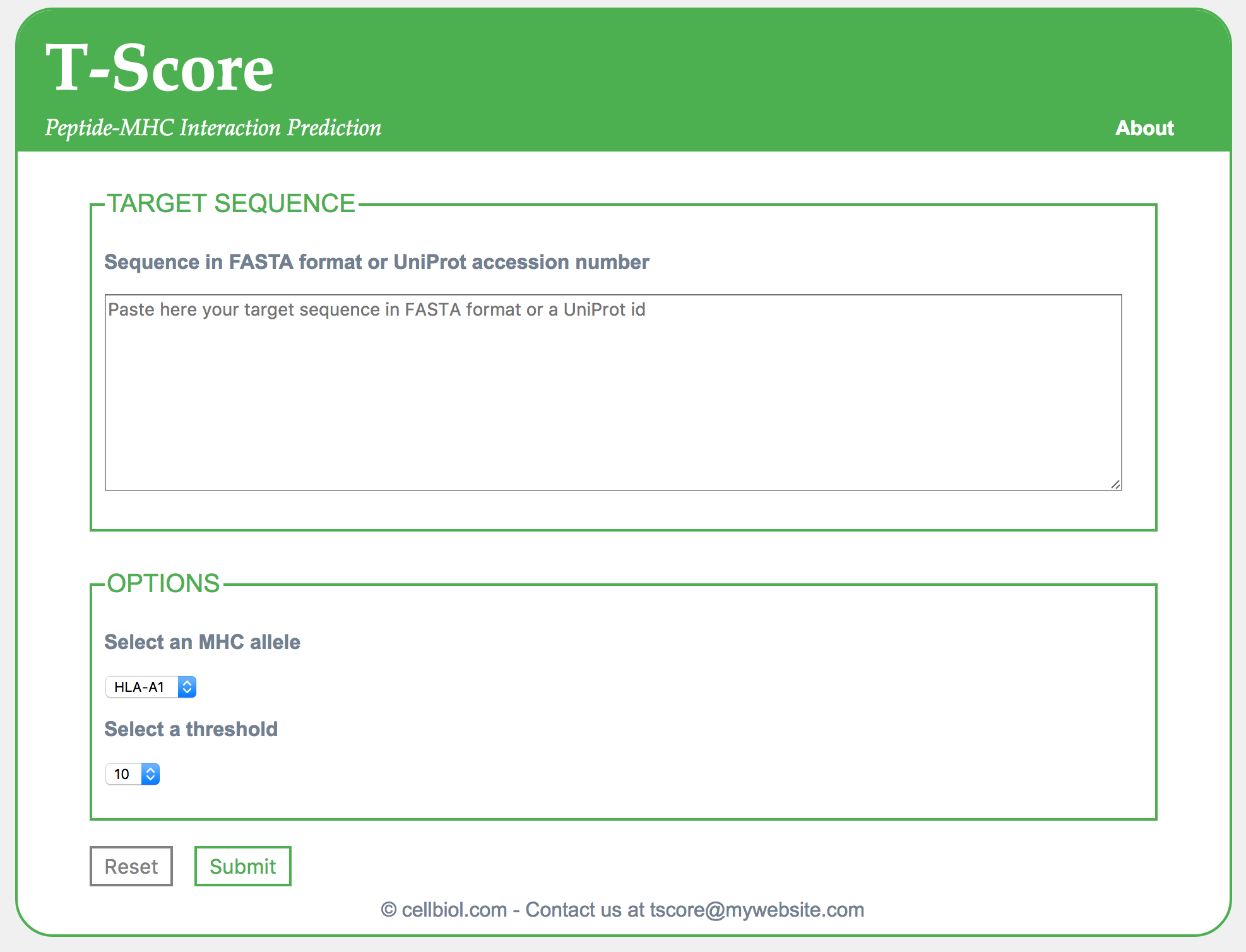
The web form will contain three fields: a text-area, a drop-down menu for MHC allele selection and a dropdown menu for selecting the threshold that we wish to apply to filter positive peptides. In the text-area, the user will be able to input either a FASTA sequence or a UniProt id. The script will then auto-detect which one of those was used as protein sequence input.
The files structure for the application will be as follows:
tscore (permission 777)
matrix (permission 777)
index.php (the web form)
script.php (the form processing script)
css
style.css
html
header.html
footer.html
include
functions.php
generate_matrices.php
DataTables download builder and CDN
As briefly mentioned in the previous section, we will use a jQuery plugin called DataTables to add a dynamic behaviour to the results table with positive peptides data presented to the user in output (more on this later). In order to do this, we do need to import jQuery and DataTables into our pages. This is done in the head part of the HTML pages by using a script tag. jQuery and DataTables also come with some dedicated stylesheets, those will have to be imported as well, through a link tag inserted in the head of our pages. There are several ways to perform those imports. For instance we could download the appropriate jQuery and DataTables scripts, and the respective CSS, locally to our web server and then import those local versions in our application web pages. There is however some complexity to this operation as we should make sure to select the appropriate versions of the scripts and import everything in the correct order for things to play smoothly.
A very convenient alternative to have a working DataTables framework in our pages is to use the DataTables download builder in combination to the DataTables Content Delivery Network (CDN). We suggest you visit those links and start to get familiar with the DataTables system. There are at least three advantages to this approach: all dependencies between the jQuery and DataTables versions used will be automatically satisfied without us having to figure them out correctly, the scripts code will be delivered fast by the cdn rather than our server and, last but not least, all the required jQuery and DataTables code can be imported with just two lines of code in the HTML, that will be provided to us by the download builder to just copy-paste in our pages source code.
The download builder provides a number of options for the DataTables system you wish or need to implement in your own pages, currently subdivided in three areas: main libraries (jQuery, type of CSS used, DataTables), extensions (various optional functionalities for the system) and packaging options (local storage or CDN delivery of the files).
A reasonable choice for the T-Score web application, in which we will use a rather simple implementation of the DataTables system, could be as follows:
Main libraries: jQuery 2.x, DataTables styling, DataTables
Extensions: FixedHeader
Packaging options: Minify, Single file, CDN
At the file of this writing, selecting those options in the download builder generates the following two lines, one for the CSS and one for the scripts, for us to include in the head of our HTTML pages:
|
1 2 3 4 |
<link rel="stylesheet" type="text/css" href="https://cdn.datatables.net/v/dt/jq-2.2.4/dt-1.10.15/fh-3.1.2/datatables.min.css"/> <script type="text/javascript" src="https://cdn.datatables.net/v/dt/jq-2.2.4/dt-1.10.15/fh-3.1.2/datatables.min.js"></script> |
Web form and CSS
We are now ready to write the code for the web form and the CSS for our pages.
header.html
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 |
<!doctype html> <html lang="en"> <head> <meta charset="utf-8"> <title>T-Score: scoring peptides and proteins for MHC binding</title> <link rel="stylesheet" type="text/css" href="https://cdn.datatables.net/v/dt/jq-2.2.4/dt-1.10.15/fh-3.1.2/datatables.min.css"/> <script type="text/javascript" src="https://cdn.datatables.net/v/dt/jq-2.2.4/dt-1.10.15/fh-3.1.2/datatables.min.js"></script> <link rel="stylesheet" href="css/style.css" type="text/css"> <script> $(document).ready(function(){ $('#results-table').dataTable({ "sDom": '<"top"i>rt<"bottom"lp><"clear">', "aoColumns": [ null, { "bSortable": false }, null, null ] }); }); </script> </head> <body> <header> <div id="header"> <div id="title-area"> <h1 id="title"><a href="index.php" id="title">T-Score</a></h1> <span id="slogan">Peptide-MHC Interaction Prediction</span> </div> <div id="navigator"> <a href="about.php" style="margin-right:10px;font-weight:bold; color:white;">About</a> </div> </div> </header> <div id="main-contents-div"> |
In the header, please note the following piece of code:
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 |
<script> $(document).ready(function(){ $('#results-table').dataTable({ "sDom": '<"top"i>rt<"bottom"lp><"clear">', "aoColumns": [ null, { "bSortable": false }, null, null ] }); }); </script> |
It runs the DataTables code on the element with id “results-table” (#results-table), the results table generated by our processing script (see script.php below).
To run DataTables with the default parameters, we could have simply used:
|
1 2 3 4 5 6 7 |
<script> $(document).ready(function(){ $('#results-table').dataTable(); }); </script> |
We did however add a little customization by using the “sDom” option, to remove the DataTables filter search box that comes by default, and the “aoColumns” option, to remove column sorting for column 2, the one with the peptide sequence, as the possibility to sort on this column does not make much sense. It makes instead a lot of sense to be able to sort the table on the first (peptide position), third (score) and third (rank) columns. A full overview of the options available for customization can be found here.
footer.html
|
1 2 3 4 5 6 7 8 |
</div> <div id="footerdiv"> © cellbiol.com - Contact us at tscore@mywebsite.com </div> </body> </html> |
index.php
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 |
<?php echo file_get_contents("html/header.html"); ?> <div id="mainformdiv"> <form action="script.php" enctype="multipart/form-data" method="post"> <div> <fieldset> <legend>TARGET SEQUENCE</legend> <p><label for="inputkin" id="inputkin-label" class="mainform-label">Sequence in FASTA format or UniProt accession number</label></p> <div class="form-field-div" id="sequence-textarea-div"> <p> <textarea name="target-sequence" placeholder="Paste here your target sequence in FASTA format or a UniProt id" class="seqinput" id="target-sequence"></textarea> </p> </div> </fieldset> </div> <fieldset style="margin-bottom: 20px;"> <legend>OPTIONS</legend> <p><label for="mhc-allele" id="mhc-allele-label" class="mainform-label">Select an MHC allele</label></p> <div class="form-field-div" id="option-div"> <p> <select id="mhc-allele" name="mhc-allele"> <option value="a1">HLA-A1</option> <option value="a2">HLA-A2</option> <option value="a3">HLA-A3</option> </select> </p> </div> <p><label for="threshold" id="threshold-label" class="mainform-label">Select a threshold</label></p> <div class="form-field-div" id="option-div"> <p> <select id="threshold" name="threshold"> <option value="10">10</option> <option value="9">9</option> <option value="8">8</option> <option value="7">7</option> <option value="6">6</option> <option value="5">5</option> <option value="4">4</option> <option value="3">3</option> <option value="2">2</option> <option value="1">1</option> </select> </p> </div> </fieldset> <input type="reset" value="Reset" class="form-reset" id="main-form-reset" style="margin-left:2px;"> <input type="submit" value="Submit" class="form-submit" id="main-form-submit" style="margin-left:2px;"> </form> <?php echo file_get_contents("html/footer.html"); ?> |
In the web form code above, we have hard-coded the select for the MHC alleles, including options for HLA-A1, A2 and A3. If we planned on a system that would accommodate for additional alleles to be eventually present in the matrix folder, we could instead generate the HTML code for the options dynamically with PHP, by looking at the .csv matrix files available in the matrix folder. The following function would be able to generate an option for each .csv file available and return the html with all the options. It supports as a single optional argument the path of the directory containing the matrices .csv files.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 |
function generate_matrix_options($matrix_dir="matrix"){ $matrix_files = scandir($matrix_dir); $available_alleles = array(); $output_html = ""; foreach($matrix_files as $matrix_file){ if(preg_match("/^(.+)-matrix\.csv$/", $matrix_file, $matches)){ $available_alleles[] = $matches[1]; } } foreach($available_alleles as $allele){ $allele_txt = "HLA-".strtoupper($allele); $output_html = $output_html."<option value=\"$allele\">$allele_txt</option>\n"; } return $output_html; } |
To use this function, we could add it to the functions.php file in the include folder. Then, in the index.php web form file, we could replace this part:
|
1 2 3 4 5 6 7 8 9 10 11 |
<div class="form-field-div" id="option-div"> <p> <select id="mhc-allele" name="mhc-allele"> <option value="a1">HLA-A1</option> <option value="a2">HLA-A2</option> <option value="a3">HLA-A3</option> </select> </p> </div> |
with this:
|
1 2 3 4 5 6 7 8 9 |
<div class="form-field-div" id="option-div"> <p> <select id="mhc-allele" name="mhc-allele"> <?php include "include/functions.php"; $options_html = generate_matrix_options(); echo $options_html ?> </select> </p> </div> |
Let’s style it.
style.css
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 101 102 103 104 105 106 107 108 109 110 111 112 113 114 115 116 117 118 119 120 121 122 123 124 125 126 127 128 129 130 131 132 133 134 135 136 137 138 139 140 141 142 143 144 145 146 147 148 149 150 151 152 153 154 155 156 157 158 159 160 161 162 163 164 165 166 167 168 169 170 171 172 173 174 175 176 177 178 179 180 181 182 183 184 185 186 187 188 189 190 191 192 193 194 195 196 197 198 199 200 |
html{ background-color:#F0F0F0; } body{ color: slategray; background-color: #FFF; /*padding: 0px 10px 10px;*/ font-family: Arial; font-size: 16px; width: 960px; height:auto; margin-right: auto; margin-left:auto; border-width: 0px; border-color: -moz-use-text-color #008000; border-radius: 30px; -moz-border-radius:30px; -webkit-border-radius:30px; border: 2px solid #4CAF50; padding-bottom: 10px; } h1{ font-size:38px; color: rgb(64,147,50); } a{ color:green; text-decoration: none; } #mainformdiv{ margin-right:auto; margin-left:auto; width:850px; } label{ cursor: pointer; font-weight:bold; } label:hover{ color:#4CAF50; } label.selected-label{ color:#4CAF50; } fieldset{ border:2px solid #4CAF50;; padding: 10px; margin-bottom: 30px; } legend{ font-size:20px; color:#4CAF50; } textarea.seqinput{ color:#2A8239; width:800px; height:150px; font-size: 14px ; } div.form-field-div{ margin-bottom: 15px; } #main-contents-div{ padding:0px 10px; } .form-reset{ -webkit-transition-duration: 0.4s; /* Safari */ transition-duration: 0.4s; cursor:pointer; margin-right: 10px; background-color: white; border: 2px solid grey; color: grey; padding: 5px 10px; text-align: center; text-decoration: none; font-size: 16px; } .form-reset:hover { background-color: grey; color: white; } .form-submit { -webkit-transition-duration: 0.4s; /* Safari */ transition-duration: 0.4s; cursor:pointer; margin-right: 10px; background-color: white; border: 2px solid #4CAF50; color: #4CAF50; padding: 5px 10px; text-align: center; text-decoration: none; font-size: 16px; } .form-submit:hover { background-color: #4CAF50; /* Green */ color: white; } #header { border-top-left-radius: 28px; border-top-right-radius: 28px; -moz-border-top-left-radius:28px; -webkit-border-top-right-radius:28px; background-color: #4CAF50;; border: 2px solid #4CAF50;; margin-bottom: 30px; } #title-area { line-height: 32px; width: 400px; /* float: left; */ display:inline-block; /* border:1px solid blue; */ padding-left:20px; } #title { text-decoration: none; color:white; font-size:52px; font-family: 'Palatino Linotype', 'Palatino', 'URW Palladio L'; -webkit-margin-before: 0.5em; -webkit-margin-after: 0.3em; /* margin-left: 20px; margin-bottom: 20px; */ /* padding-bottom: 0px; padding-top: 0; */ } #navigator{ width: 500px; display:inline-block; text-align:right; vertical-align:middle; /* border:1px solid red; */ } #navigator li{ display:inline; margin-left:20px; } #navigator a{ color:white; } #slogan { font-family: 'Palatino Linotype', 'Palatino', 'URW Palladio L'; font-style: italic; color: white; font-size: 18px; text-align: left; } .sequence{ font-family:courier; } #results-div{ width:800px; margin-left: auto; margin-right: auto; } th{ text-align: left; } #footerdiv{ text-align:center; margin-top:20px; } |
Data processing
For the functions.php file please refer to the previous section, as it remains unchanged. It should contain the following functions:
– process_fasta()
– pepscore()
– matrix_file_to_array()
– slide_window()
– generate_matrix_options() (optional, discussed above in this section)
script.php
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 |
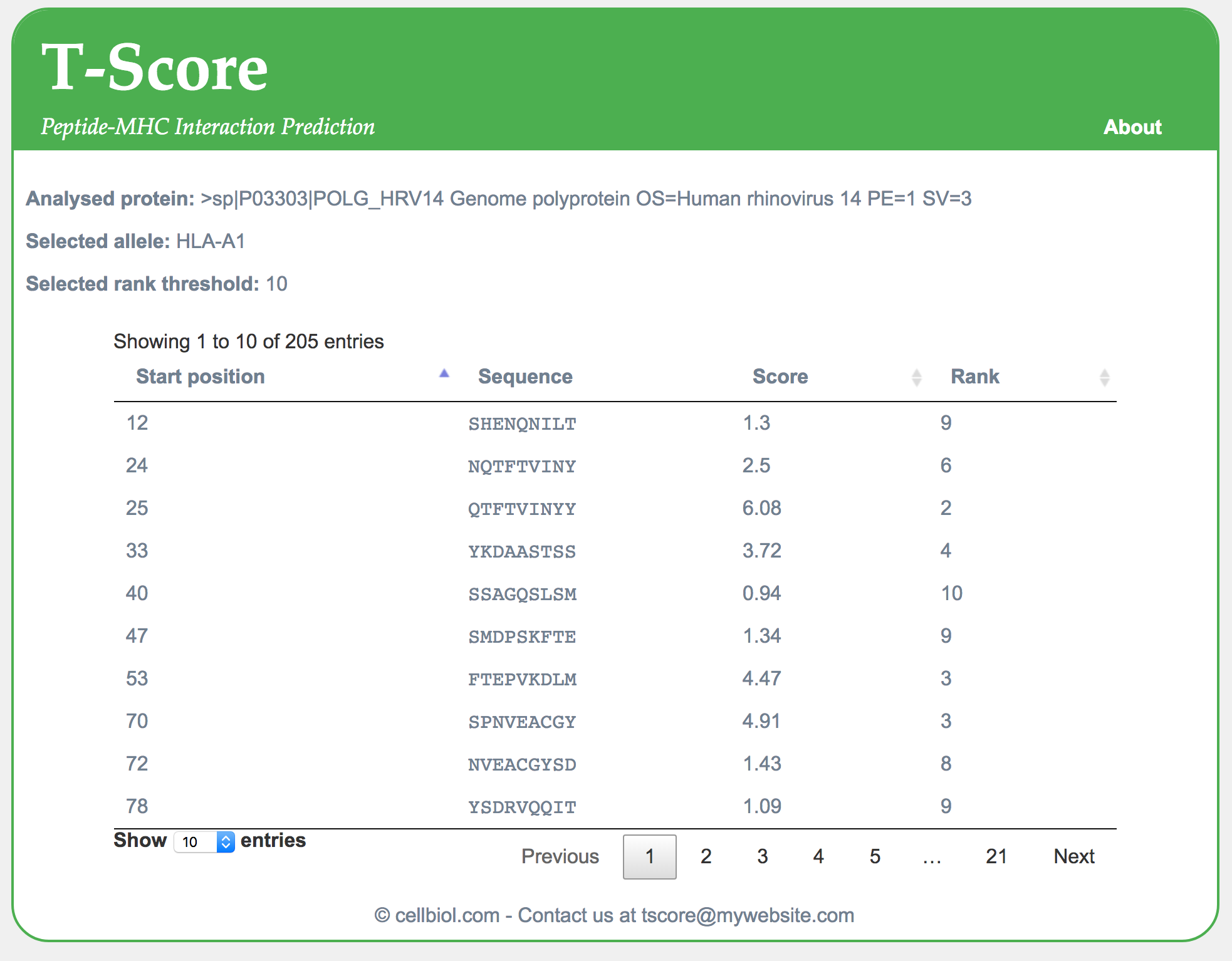
<?php include "include/functions.php"; $target_sequence = trim($_POST["target-sequence"]); $mhc_allele = $_POST["mhc-allele"]; $threshold = $_POST["threshold"]; $mhc_allele_txt = "HLA-".strtoupper($mhc_allele); $html_header = file_get_contents("html/header.html"); $html_footer = file_get_contents("html/footer.html"); if($target_sequence == ""){ // We really need a target sequence to proceed die("$html_header<p>Target sequence information missing, please <a href=\"index.php\">Go back</a> and try again</p>$html_footer"); } if(!preg_match("/>/", $target_sequence)){ // If the sequence field does not contain a > symbol, we assume it is a UniProt id and try to retrieve it from uniprot, as already discussed in section 4-6 $fasta_sequence = file_get_contents("http://www.uniprot.org/uniprot/".$target_sequence.".fasta"); // If this fails, UniProt will issue an error HTML page that has a DOCTYPE in the first line. We use this to detect failures and stop the script from going further if(preg_match("/DOCTYPE/", $fasta_sequence)){ die("$html_header<p>The target sequence information is not a FASTA sequence, so it was assumed to be a UniProt ID that however could not be retrieved from UniProt. Please <a href=\"index.php\">Go back</a>, check your data and try again</p>$html_footer"); } } else{ // If the target sequence field contains a > it is assumed to be a FASTA sequence $fasta_sequence = $target_sequence; } // At this point we should have a valid sequence in FASTA format, stored in the $fasta_sequence variable, ready to be processed further $process_fasta = process_fasta($fasta_sequence); $sequence = $process_fasta[1]; $seq_header = $process_fasta[0]; $peptides = slide_window($sequence); $matrix = matrix_file_to_array("matrix/$mhc_allele-matrix.csv"); // MHC allele values can be a1, a2 or a3. We embed this in a string to reflect the path of the corresponding .csv file and get the matrix // We now use the code already written in the previous section to compute positive peptides with their scores and ranks $results = array(); // This is where we store all the data for peptides that rank at least 10 or below, "positive" peptides // Remember that the lower the rank is, the best binder a peptide is predicted to be. The other peptides, the "negatives", will be discarded and not shown in the output $pos_counter = 1; // To keep track of the start position of each peptide in the sequence foreach($peptides as $peptide){ $pepscore = pepscore($peptide, $matrix); $score = $pepscore[0]; $rank = $pepscore[1]; if($rank <= $threshold){ $results[] = array($pos_counter, $peptide, $score, $rank); // In the results array each peptide is represented by an array storing the peptide sequence, start position, score and rank } $pos_counter++; // The position counter is incremented by one each time we proceed to the next peptide } $total_peptides_num = $pos_counter; $positive_peptides_num = count($results); echo $html_header; echo "<p><strong>Analysed protein:</strong> </p><p>$seq_header</p>\n"; echo "<p><strong>Selected allele:</strong> $mhc_allele_txt</p>\n"; echo "<p><strong>Selected rank threshold:</strong> $threshold</p>\n"; echo "<style>td{padding:5px;}\nthead{font-weight:bold}</style>\n"; echo "<table id=\"results-table\">\n<thead>\n<tr><th>Start position</th><th>Sequence</th><th>Score</th><th>Rank</th></tr>\n</thead>\n<tbody>\n"; foreach($results as $result){ $start = $result[0]; $end = $start + 8; $pepseq = $result[1]; $pepscore = $result[2]; $peprank = $result[3]; echo "<tr><td>$start</td><td><span class=\"sequence\">$pepseq</span></td><td>$pepscore</td><td>$peprank</td></tr>\n"; } echo "</tbody>\n</table>"; echo $html_footer; ?> |
You can test the script live here. Find a screenshot of the output with protein UniProt P03303 below.
Chapter Sections
[pagelist include=”1461″]
[siblings]
WORK IN PROGRESS ON CHAPTER 5!