In this section we will build a web application able to color amino-acids in a protein sequence according to their nonpolar, polar, basic or acidic nature (see figure 4-7-2). The application will provide some simple statistics on the protein sequence composition in regard to those classes of amino-acids. These data will be represented graphically by building an HTML based graph.
The web form will be very simple, with just a text-area for the sequence field and a submit button.
The PHP code to handle the sequence for amino-acids classification will be based on the amino-acids classification script we saw in section 4-7.
The script will support sequences in FASTA format. To handle FASTA sequences we will use the process_fasta() function that we wrote in section 4-12.
This is what the output of the script will look like:
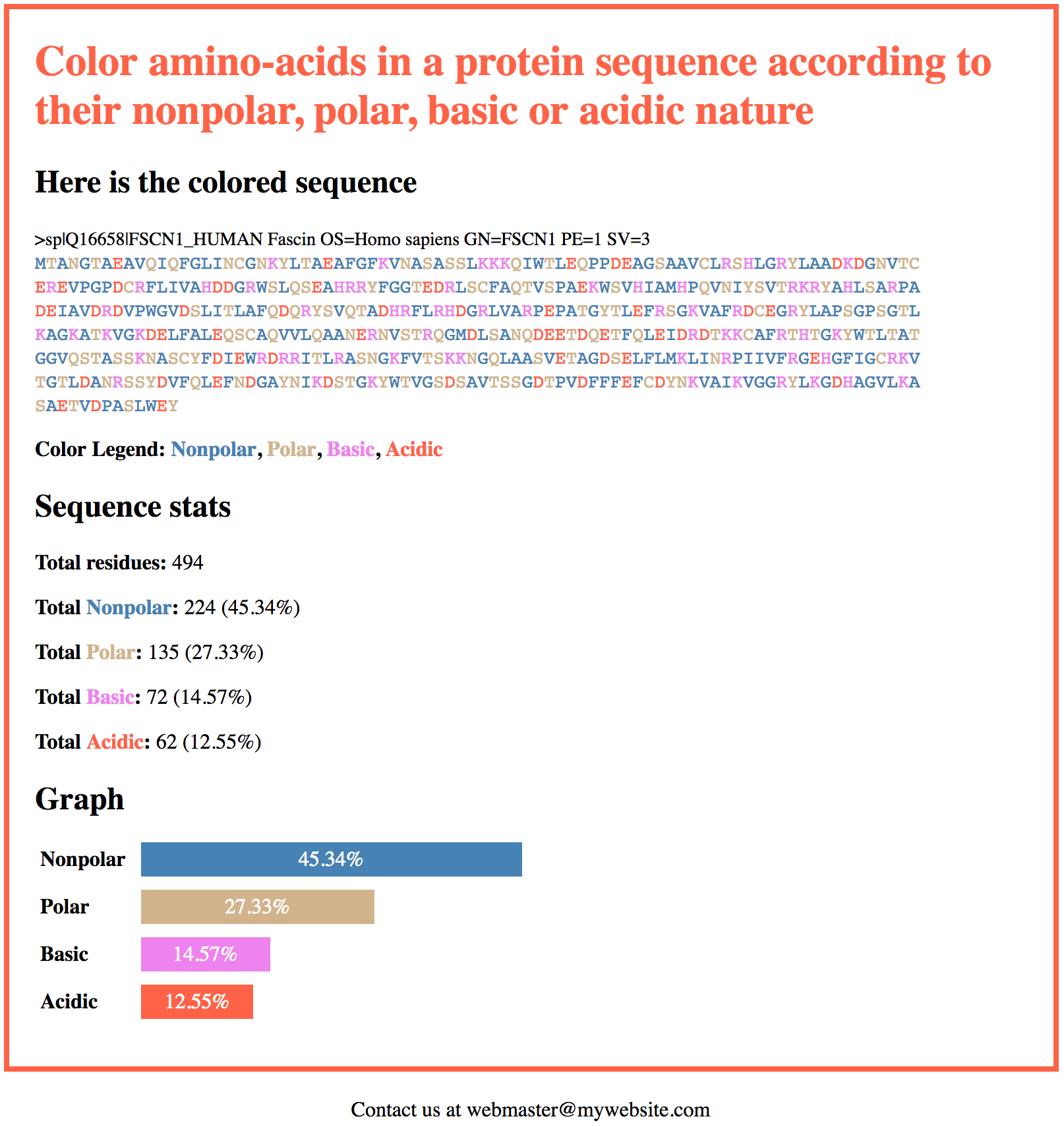
Coloring amino-acids
In order to color amino-acids in the script output we embed each amino-acid letter in a span tag with the appropriate class “nonpolar”, “polar”, “basic” or “acidic”:
|
1 2 3 |
<span class="acidic">D</span> |
In the stylesheet we assign a color to the class:
|
1 2 3 4 5 |
.acidic{ color:tomato; } |
Counting amino-acids
To count the amino-acids in each class we define four counters, one for each class, before we start cycling through the amino-acids array. We also use an $i counter to count the total number of residues and keep track of where we stand during the cycle. $i starts at 1 while the “classes counters” start at 0.
|
1 2 3 4 5 6 7 8 |
$i = 1; // to keep track of the amino-acid number in the cycle $num_nonpolar = 0; // Number of nonpolar residues $num_polar = 0; // Number of polar residues $num_basic = 0; // Number of basic residues $num_acidic = 0; // Number of acidic residues $num_undef = 0; // Number of undefined residues |
Each time we find an amino-acid belonging to a particular class during the foreach cycle that iterates through the amino-acids array, the respective counter is incremented by one:
|
1 2 3 4 5 6 |
if(strrchr($nonpolar, $aminoacid)){ ..... $num_nonpolar++; } |
Once we have the total number of amino-acids and the number for each class, we can calculate percentages:
|
1 2 3 |
$percent_nonpolar = round(($num_nonpolar*100)/$i, 2); |
We limit the digits of the float number resulting from the division in the calculation by using the PHP round() function, that as the name implies, can round the float to a defined number of digits. The round() function takes the float as first argument and the number of desired digits as second argument. If no second argument is provided, the default of zero digits is used (the float is rounded to an integer).
Building a graph in HTML
To build a graph in HTML we use a description list, the dl tag.
The general syntax of a description list is as follows. In this example we have a description list with 3 items (Item 1, Item 2, Item 3) and their respective descriptions (Description 1, Description 2, Description 3):
|
1 2 3 4 5 6 7 8 9 10 |
<dl> <dt>Item 1</dt> <dd>Description 1</dd> <dt>Item 2</dt> <dd>Description 2</dd> <dt>Item 3</dt> <dd>Description 3</dd> </dl> |
This code as it is will yield the following (what you see below may be influenced by this website’s style sheet, try it on a page yourself):
- Item 1
- Description 1
- Item 2
- Description 2
- Item 3
- Description 3
In our graph, the items are the names of the amino-acids classes, that are set to float:left in the CSS, and the descriptions contain divs. The script sets the width attribute of each div as the percentage of amino-acids in the corresponding class. The background-color attribute of each div is set to the same color we have selected for the amino-acids class by assigning to the div a “bar-nonpolar”, “bar-polar”, “bar-basic” or “bar-acidic” class, check out the CSS stylesheet below.
This is an example of the resulting markup with a sample sequence. Again, mind that for the graph to look like the graph we get by running the script, CSS definitions for the various elements (dl, dd, dt and the bar classes assigned to the divs) are essential:
|
1 2 3 4 5 6 7 8 9 10 11 12 |
<dl> <dt>Nonpolar</dt> <dd><div class="bar bar-nonpolar" style="width:45.34%">45.34%</div></dd> <dt>Polar</dt> <dd><div class="bar bar-polar" style="width:27.33%">27.33%</div></dd> <dt>Basic</dt> <dd><div class="bar bar-basic" style="width:14.57%">14.57%</div></dd> <dt>Acidic</dt> <dd><div class="bar bar-acidic" style="width:12.55%">12.55%</div></dd> </dl> |
The script code
As for all the web applications in this book, the code for this application will be distributed across several files. The files structure will be similar to the one used for the code in the previous section, with the addition of an include folder containing a functions.php file where the process_fasta() function will be stored.
color_sequence
index.php
script.php
html
header.html
footer.html
css
style.css
include
functions.php
header.html
|
1 2 3 4 5 6 7 8 9 10 11 12 |
<!DOCTYPE html> <html lang="en"> <head> <meta charset="UTF-8"> <title>A script to color a protein sequence</title> <link rel="stylesheet" href="css/style.css" type="text/css"> </head> <body> <div id="main-contents"> <h1>Color amino-acids in a protein sequence according to their nonpolar, polar, basic or acidic nature</h1> |
footer.html
|
1 2 3 4 5 6 7 8 |
</div> <footer> Contact us at webmaster@mywebsite.com </footer> </body> </html> |
index.php
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 |
<?php echo file_get_contents("html/header.html"); ?> <form action="script.php" method="POST" enctype="multipart/form-data" id="revcomp_form"> <fieldset id="data"> <legend>Data</legend> <p> <label for="fasta_sequence">Your input sequence - FASTA format supported</label><br> <textarea name="fasta_sequence" id="fasta_sequence"></textarea> </p> </fieldset> <input type="submit" value="Color Sequence"> </form> <?php echo file_get_contents("html/footer.html"); ?> |
In the CSS file, with respect to the example in the previous section, we add four classes (nonpolar, polar, basic, acidic) for the four types of amino-acids and assign a different color to each class. We add an “undefined” class, for unexpected characters in the sequence, and a “sequence” class with font-family:courier that we will apply to the whole output sequence. We also add some styles for the description list elements (dl, dt and dd) and the divs (“bar” classes) used to generate the graph.
For more advanced graph generation with description list tags and CSS check out this tutorial on htmlgoodies.com.
For an uber-cool dynamic graph generated with javascript and the HTML5 canvas element, check out this page on the williammalone.com web site.
style.css
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 101 102 103 104 105 106 107 108 109 110 111 112 113 114 115 116 117 118 119 120 121 122 123 124 125 126 127 128 129 130 131 132 133 134 135 136 137 138 139 |
body{ width: 800px; margin-right:auto; margin-left:auto; } #main-contents{ border:4px solid tomato; margin-right:auto; margin-left:auto; margin-bottom: 20px; padding:20px; padding-top:0; } .bar { margin-bottom: 10px; color: #fff; padding: 4px; text-align: center; } dl{ width:700px; } dt{ float: left; padding: 4px; font-weight:bold; } dd{ margin-left:80px; } span.sequence{ font-family:courier; font-size:14px; } span.header{ font-size:14px; } .nonpolar{ color:SteelBlue; } .bar-nonpolar{ background-color:SteelBlue; } .polar{ color:tan; } .bar-polar{ background-color:tan; } .basic{ color:violet; } .bar-basic{ background-color:violet; } .acidic{ color:tomato; } .bar-acidic{ background-color:tomato; } span.undefined{ color:darkgrey; } label{ cursor:pointer; font-weight:bold; color:teal; } label.radio{ font-weight:normal; } span.field-title{ font-weight:bold; color:teal; } fieldset{ border:1px solid tomato; margin-bottom:20px; } legend{ font-weight:bold; color:tomato; } input[type=submit] { background-color: tomato; border: none; color: white; padding: 5px 10px; text-decoration: none; margin: 4px 2px; cursor: pointer; text-transform: uppercase; font-weight:bold; } h1{ color:tomato; } textarea{ width:500px; height:200px; font-family:courier; background:whitesmoke; } input[type="text"]{ background:whitesmoke; } footer{ text-align:center; } |
functions.php
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 |
<?php function process_fasta($fasta_sequence, $mode="all"){ $fasta_lines = explode("\n", $fasta_sequence); $header = "> Generic"; // We will store the header line here during the next foreach cycle $sequence = ""; // We will store the sequence here during the next foreach cycle foreach($fasta_lines as $line){ // We strip possible whitespace (or other characters) from the beginning and end of the line $line = trim($line); if(preg_match("/^>/", $line)){ // If the line starts with a > it's the header line $header = $line; } elseif($line != ""){ $sequence = $sequence.$line; // We concatenate each new sequence line in the $sequence variable } } // At this point we should have the FASTA header in the $header variable // and the whole sequence in the $sequence variable // And now the return part, that depends on value of $mode if($mode == "all"){ return array($header, $sequence); } elseif($mode == "seq"){ return $sequence; } elseif($mode == "header"){ return $header; } else{ return "WARNING: process_fasta mode not supported"; } } ?> |
script.php
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 101 102 103 104 105 106 107 108 109 110 111 112 113 114 115 116 117 118 119 120 121 122 123 124 125 |
<?php include("include/functions.php"); // We include the functions.php file $fasta_sequence = $_POST["fasta_sequence"]; // and grab the sequence submitted by the user through the web form // We call it fasta_sequence rather than sequence as it may well be a FASTA sequence. The name of the // field in the web form (index.php) was also changed accordingly $sequence = strtoupper(process_fasta($fasta_sequence, "seq")); // We use process_fasta() to extract the "raw" sequence // from the fasta_sequence and also make sure it is in uppercase characters $header = process_fasta($fasta_sequence, "header"); // We use process_fasta() to extract the header $aminoacids = str_split($sequence, 1); // Generating an array with the amino-acids as elements $out_sequence = "<span class=\"sequence\">"; // This string will contain the colored sequence to output to the web page // For now it only contains the opening span tag for the sequence with a class "sequence" that has // a font-family:courier as defined in the style.css file $unexpected_chars = array(); // In case we find some unexpected character in the sequence, we will store it here to keep track // We define four strings containing the different classes of amino-acids $nonpolar ="FLIMVPAWG"; // A string made by all the nonpolar amino-acids in single letter notation $polar = "STYCQN"; // Polar amino-acids $basic = "HKR"; // Basic amino-acids $acidic = "DE"; // Acidic amino-acids // We now iterate through the amino-acids in the input sequence (the $aminoacids array) // For each amino-acid, we check if it is nonpolar, polar, basic or acidic // and wrap it in a span tag with the correct class. This, together with the CSS declarations in the style.css file, // will color the amino-acid in the output web page // Every 80 amino-acids we add a break tag, to respect the FASTA format $i = 1; // to keep track of the amino-acid number in the cycle $num_nonpolar = 0; // Number of nonpolar residues $num_polar = 0; // Number of polar residues $num_basic = 0; // Number of basic residues $num_acidic = 0; // Number of acidic residues $num_undef = 0; // Number of undefined residues foreach($aminoacids as $aminoacid){ if(strrchr($nonpolar, $aminoacid)){ $out_sequence = $out_sequence."<span class=\"nonpolar\">$aminoacid</span>"; $num_nonpolar++; } elseif(strrchr($polar, $aminoacid)){ $out_sequence = $out_sequence."<span class=\"polar\">$aminoacid</span>"; $num_polar++; } elseif(strrchr($basic, $aminoacid)){ $out_sequence = $out_sequence."<span class=\"basic\">$aminoacid</span>"; $num_basic++; } elseif(strrchr($acidic, $aminoacid)){ $out_sequence = $out_sequence."<span class=\"acidic\">$aminoacid</span>"; $num_acidic++; } else{ // We leave the possibility open to encounter an unknown character we have not classified $out_sequence = $out_sequence."<span class=\"undefined\">$aminoacid</span>"; $num_undef++; $unexpected_chars[] = array($aminoacid, $i); } if(is_int($i/80)){ // Every 80 amino-acids we add a break (and a newline to make the source code of the web page more readable) $out_sequence = $out_sequence."<br>\n"; } $i++; } $out_sequence = $out_sequence."</span>\n"; // We close the sequence span tag // Calculating percentages // The round() function rounds a float (first argument) to the number of decimals we specify as second argument $percent_nonpolar = round(($num_nonpolar*100)/$i, 2); $percent_polar = round(($num_polar*100)/$i, 2); $percent_basic = round(($num_basic*100)/$i, 2); $percent_acidic = round(($num_acidic*100)/$i, 2); // Providing an output to the user // We embed the output data within the same header and footer HTML code used in the web form // to ensure a consistent navigation experience and provide the feel that // everything takes place "in the same website" echo file_get_contents("html/header.html"); // Writing the header HTML to the output page echo "<h2>Here is the colored sequence</h2>\n"; echo "<p><span class=\"header\">$header</span><br>\n"; echo "$out_sequence</p>\n"; echo "<p><strong>Color Legend: <span class=\"nonpolar\">Nonpolar</span>, <span class=\"polar\">Polar</span>, <span class=\"basic\">Basic</span>, <span class=\"acidic\">Acidic</span></strong>"; echo "<h2>Sequence stats</h2>"; echo "<p><strong>Total residues: </strong>$i</p>"; echo "<p><strong>Total <span class=\"nonpolar\">Nonpolar</span>: </strong>$num_nonpolar ($percent_nonpolar%)</p>"; echo "<p><strong>Total <span class=\"polar\">Polar</span>: </strong>$num_polar ($percent_polar%)</p>"; echo "<p><strong>Total <span class=\"basic\">Basic</span>: </strong>$num_basic ($percent_basic%)</p>"; echo "<p><strong>Total <span class=\"acidic\">Acidic</span>: </strong>$num_acidic ($percent_acidic%)</p>"; // We generate a graph by using a description list dl tag // with div elements in the dd tags // To get a nice graph from this markup the CSS is very important, // check out the CSS definitions for dl, dd, dt and the bar classes in the style.css file above echo "<h2>Graph</h2>"; echo "<p><dl> <dt>Nonpolar</dt> <dd><div class=\"bar bar-nonpolar\" style=\"width:$percent_nonpolar%\">$percent_nonpolar%</div></dd> <dt>Polar</dt> <dd><div class=\"bar bar-polar\" style=\"width:$percent_polar%\">$percent_polar%</div></dd> <dt>Basic</dt> <dd><div class=\"bar bar-basic\" style=\"width:$percent_basic%\">$percent_basic%</div></dd> <dt>Acidic</dt> <dd><div class=\"bar bar-acidic\" style=\"width:$percent_acidic%\">$percent_acidic%</div></dd> </dl></p>"; if($num_undef != 0){ // If we have any unexpected character echo "<p><strong>The following $num_undef unexpected characters were found:</strong><br>\n"; foreach($unexpected_chars as $char_arr){ $char = $char_arr[0]; $pos = $char_arr[1]; echo "$char...$pos<br>\n"; } echo "</p>"; } echo file_get_contents("html/footer.html"); // Writing the footer HTML to the output page ?> |
You may test the script live here.
Chapter Sections
[pagelist include=”1461″]
[siblings]
WORK IN PROGRESS ON CHAPTER 5!