In the previous section we have started to see some basics on how to manipulate strings and biological sequences in PHP by using predefined functions. In this section we explore the topic further by exploring a few more useful PHP built-in tools (predefined functions).
Splitting a biological sequence in single nucleotides, codons or amino-acids with the str_split() function
str_split() – string split – requires two arguments, a string and a number (an integer). It allows, as the name suggests, to split a string in pieces composed by a certain number (the second argument passed on function call) of characters. If the passed number exceeds the string length, it will return the entire string. If the string length cannot be exactly divided by the number, it will return sub strings formed by the passed number and a last substring with what remains.
Let’s make this more clear with a few examples.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 |
<?php $mystring = "123456789"; $splitted_on_3 = str_split($mystring,3); // splitting to sub-strings or 3 characters echo "<p>Splitting 123456789 to substrings of 3 characters<br>"; var_dump($splitted_on_3); echo "</p>"; // Will output: array(3) { [0]=> string(3) "123" [1]=> string(3) "456" [2]=> string(3) "789" } $splitted_on_2 = str_split($mystring,2); // splitting to sub-strings or 2 characters echo "<p>Splitting 123456789 to substrings of 2 characters<br>"; var_dump($splitted_on_2); // Will output: array(5) { [0]=> string(2) "12" [1]=> string(2) "34" [2]=> string(2) "56" [3]=> string(2) "78" [4]=> string(1) "9" } echo "</p>"; $splitted_on_10 = str_split($mystring,10); // splitting to sub-strings or 10 characters echo "<p>Splitting 123456789 to substrings of 10 characters<br>"; var_dump($splitted_on_10); // Will output: array(1) { [0]=> string(9) "123456789" } echo "</p>"; ?> |
In the example above please note that when we try to split a 9 characters string in substrings of 2 characters, str_split() will generate an array of 5 elements. The first 4 are composed by substrings of 2 characters (what we asked for by passing 2 as second argument to the function) and the last by just one character, that constitutes the remainder of the string after having taken out all the possible two characters substrings.
Also note that when we attempt to subdivide our string in substrings longer than the string itself – in this example we try to subdivide a 9 characters string in substrings of 10 – the whole string is returned as the single element of the str_split() output array.
In the example that follows we use str_split() to subdivide a coding sequence into the codons (triplets) that compose it. You may see as this could be a first step toward a translation of our DNA coding sequence into a protein sequence.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 |
<?php $cod_sequence = "ATGGCTAATGATAGA"; // A short portion of a DNA coding sequence $codons = str_split($cod_sequence,3); echo "<p>\n<strong>Here are the codons composing $cod_sequence</strong>\n<ul>\n"; foreach($codons as $codon){ echo "<li>".$codon."</li>\n"; } echo "</ul>\n</p>"; ?> |
This will generate the following output:
Here are the codons composing ATGGCTAATGATAGA
- ATG
- GCT
- AAT
- GAT
- AGA
If we want to split the same DNA sequence used in the previous example into single nucleotides instead codons, all we have to do is use 1 instead of 3 as an argument in the str_split() call. Let’s also change the variables names so that they make sense for the new script.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 |
<?php $cod_sequence = "ATGGCTAATGATAGA"; // A short portion of a DNA coding sequence $nucleotides = str_split($cod_sequence,1); echo "<p>\n<strong>Here are the nucleotides composing $cod_sequence</strong>\n<ul>\n"; foreach($nucleotides as $nucleotide){ echo "<li>".$nucleotide."</li>\n"; } echo "</ul>\n</p>"; ?> |
Here is the output of the script above:
Here are the nucleotides composing ATGGCTAATGATAGA
- A
- T
- G
- G
- C
- T
- A
- A
- T
- G
- A
- T
- A
- G
- A
How to reverse-complement a DNA sequence in PHP
We now have enough knowledge of PHP to perform a simple and basic, yet often essential operation that concerns DNA sequences: from one strand, extrapolate the other. You are surely familiar with the concept that DNA is a double helix and the two strands of the helix are complementary to each other: if A is on one strand, T is on the other (and vice-versa) and if C is on one strand G is on the other (and vice-versa).
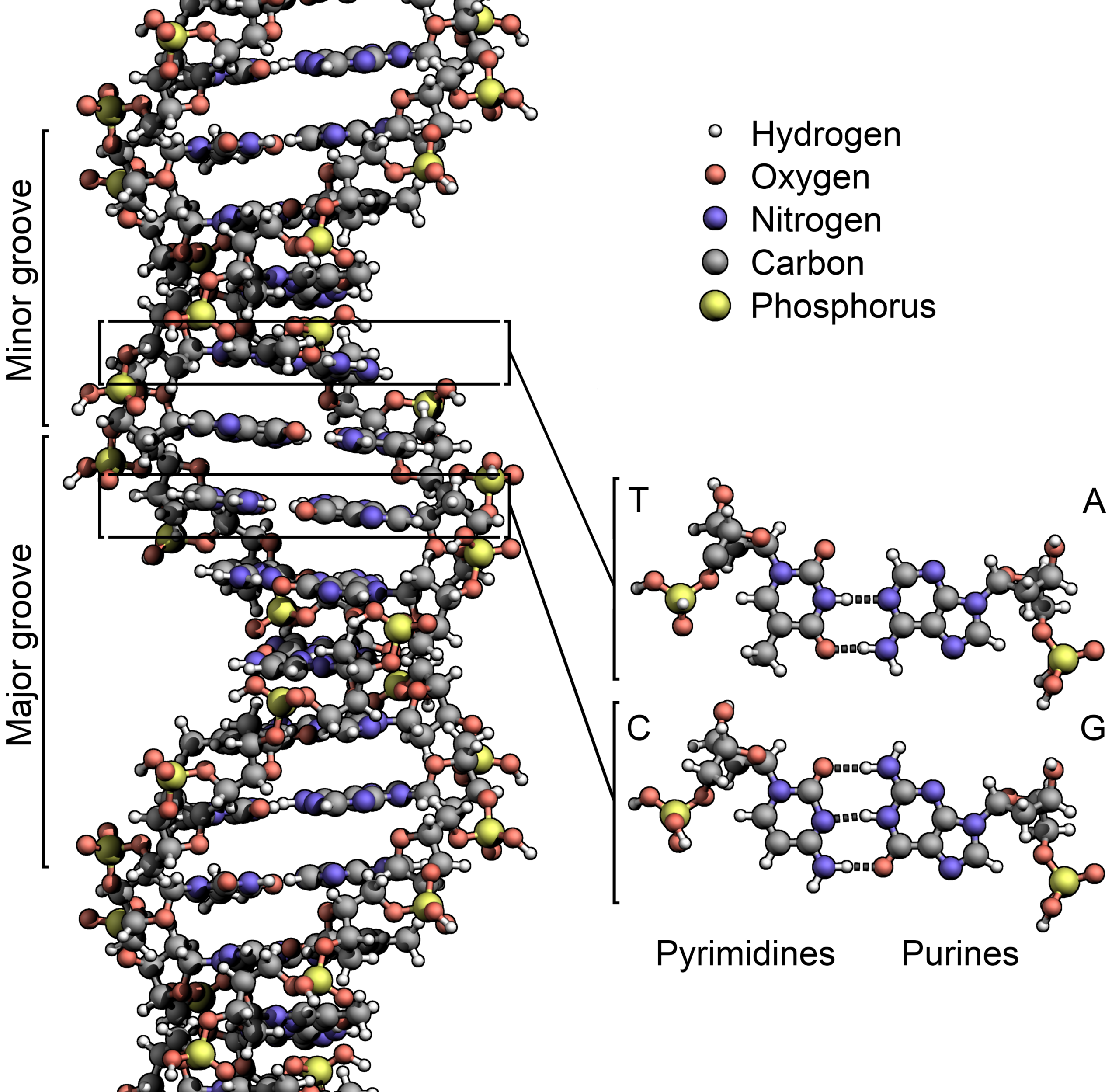
Given the sequence of one DNA strand, you can easily obtain the sequence of the other by performing a so called “reverse complement” operation.
Here is some PHP code that allows you to perform just that. We will explore this much further on the web applications chapter.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 |
<?php $complement_dict = array( "A" => "T", "T" => "A", "G" => "C", "C" => "G" ); $sequence = "ATGGTGAAGCAGATCGA"; $nucleotides = str_split($sequence,1); $complement_sequence = ""; foreach($nucleotides as $nucleotide){ $complement_sequence = $complement_sequence.$complement_dict[$nucleotide]; } $revcomp_sequence = strrev($complement_sequence); echo "<p>\n<strong>Input Sequence</strong><br>\n<span style=\"font-family:courier;\">$sequence</span>\n</p>\n<p>\n<strong>Reverse Complement</strong><br>\n<span style=\"font-family:courier;\">$revcomp_sequence</span>\n</p>"; ?> |
Here is the output of this script:
Input Sequence
ATGGTGAAGCAGATCGA
Reverse Complement
TCGATCTGCTTCACCAT
Translating a DNA coding sequence to an amino-acids sequence with PHP
Let’s now take the splitting of a DNA sequence into codons shown above one step further and actually perform the translation of the DNA coding sequence to an amino-acids sequence. In order to do a translation, pretty much any translation from a language to another, we do need a dictionary where we can look for a word and get the translated word for the new language we are interested in. In the case of DNA codons, we need a dictionary to translate triplets of DNA nucleotides (codons) to the correspond amino-acids. We can easily generate such a dictionary in PHP from the genetic code.
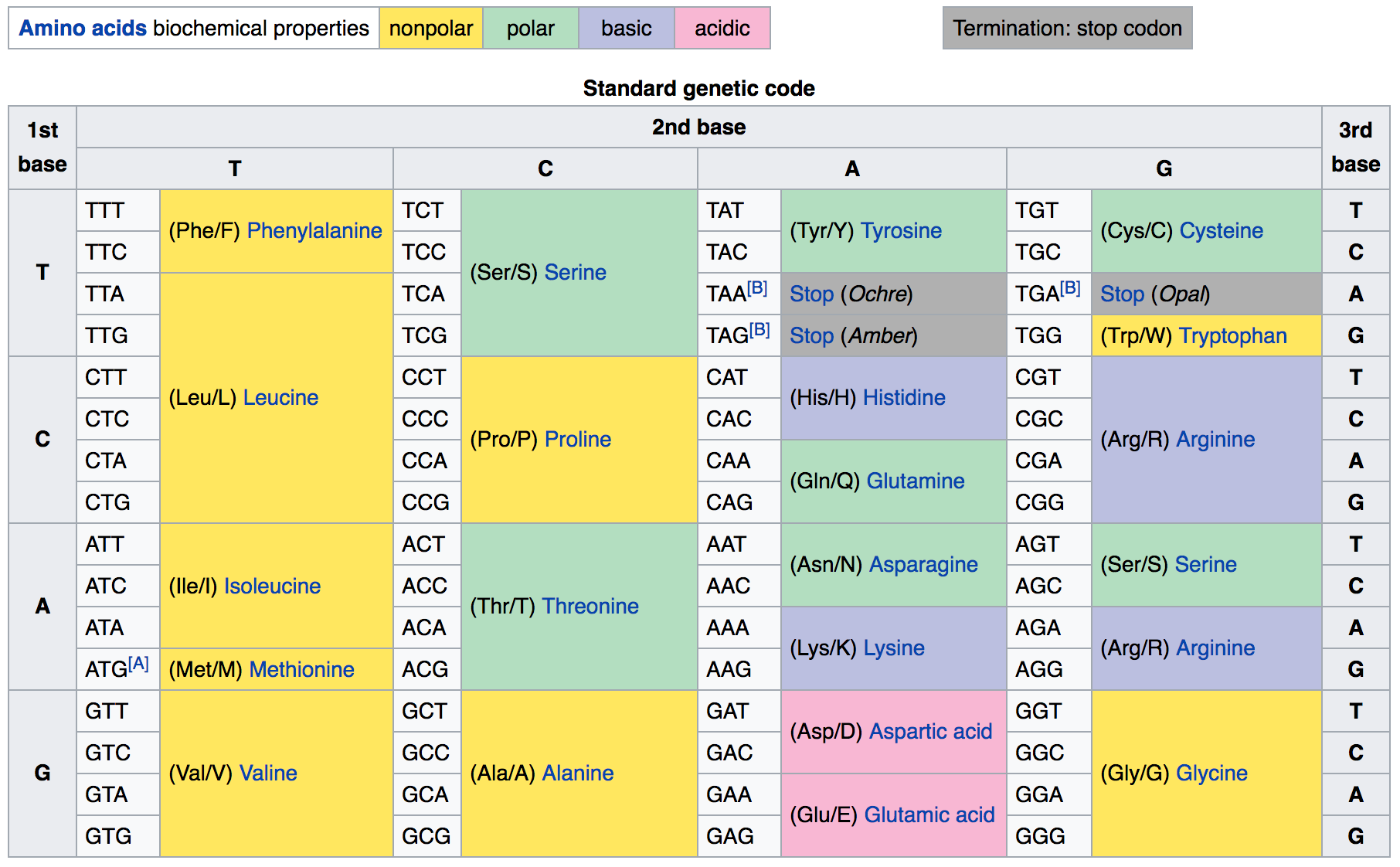
As you know at this point, a dictionary can be created in PHP as an array in which each key is associated to a value. Each key is the word to translate while the corresponding value is the translation itself.
Let’s get into it. Here’s the genetic code as a PHP array dictionary, derived from the figure above:
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 |
<?php $genetic_code = array( "TTT" => "F", "TTC" => "F", "TTA" => "L", "TTG" => "L", "CTT" => "L", "CTC" => "L", "CTA" => "L", "CTG" => "L", "ATT" => "I", "ATC" => "I", "ATA" => "I", "ATG" => "M", "GTT" => "V", "GTC" => "V", "GTA" => "V", "GTG" => "V", "TCT" => "S", "TCC" => "S", "TCA" => "S", "TCG" => "S", "CCT" => "P", "CCC" => "P", "CCA" => "P", "CCG" => "P", "ACT" => "T", "ACC" => "T", "ACA" => "T", "ACG" => "T", "GCT" => "A", "GCC" => "A", "GCA" => "A", "GCG" => "A", "TAT" => "Y", "TAC" => "Y", "TAA" => "Stop", "TAG" => "Stop", "CAT" => "H", "CAC" => "H", "CAA" => "Q", "CAG" => "Q", "AAT" => "N", "AAC" => "N", "AAA" => "K", "AAG" => "K", "GAT" => "D", "GAC" => "D", "GAA" => "E", "GAG" => "E", "TGT" => "C", "TGC" => "C", "TGA" => "Stop", "TGG" => "W", "CGT" => "R", "CGC" => "R", "CGA" => "R", "CGG" => "R", "AGT" => "S", "AGC" => "S", "AGA" => "R", "AGG" => "R", "GGT" => "G", "GGC" => "G", "GGA" => "G", "GGG" => "G" ); ?> |
We will use this genetic code PHP dictionary to translate an actual DNA coding sequence. Let’s take the Human Thioredoxin (Uniprot P10599) as an example. The coding sequence can be found here.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 |
<?php $genetic_code = array( "TTT" => "F", "TTC" => "F", "TTA" => "L", "TTG" => "L", "CTT" => "L", "CTC" => "L", "CTA" => "L", "CTG" => "L", "ATT" => "I", "ATC" => "I", "ATA" => "I", "ATG" => "M", "GTT" => "V", "GTC" => "V", "GTA" => "V", "GTG" => "V", "TCT" => "S", "TCC" => "S", "TCA" => "S", "TCG" => "S", "CCT" => "P", "CCC" => "P", "CCA" => "P", "CCG" => "P", "ACT" => "T", "ACC" => "T", "ACA" => "T", "ACG" => "T", "GCT" => "A", "GCC" => "A", "GCA" => "A", "GCG" => "A", "TAT" => "Y", "TAC" => "Y", "TAA" => "Stop", "TAG" => "Stop", "CAT" => "H", "CAC" => "H", "CAA" => "Q", "CAG" => "Q", "AAT" => "N", "AAC" => "N", "AAA" => "K", "AAG" => "K", "GAT" => "D", "GAC" => "D", "GAA" => "E", "GAG" => "E", "TGT" => "C", "TGC" => "C", "TGA" => "Stop", "TGG" => "W", "CGT" => "R", "CGC" => "R", "CGA" => "R", "CGG" => "R", "AGT" => "S", "AGC" => "S", "AGA" => "R", "AGG" => "R", "GGT" => "G", "GGC" => "G", "GGA" => "G", "GGG" => "G" ); $translated_sequence = ""; $thio_human_cds = "ATGGTGAAGCAGATCGAGAGCAAGACTGCTTTTCAGGAAGCCTTGGACGCTGCAGGTGATAAACTTGTAGTAGTTGACTTCTCAGCCACGTGGTGTGGGCCTTGCAAAATGATCAAGCCTTTCTTTCATTCCCTCTCTGAAAAGTATTCCAACGTGATATTCCTTGAAGTAGATGTGGATGACTGTCAGGATGTTGCTTCAGAGTGTGAAGTCAAATGCATGCCAACATTCCAGTTTTTTAAGAAGGGACAAAAGGTGGGTGAATTTTCTGGAGCCAATAAGGAAAAGCTTGAAGCCACCATTAATGAATTAGTCTAA"; $codons = str_split($thio_human_cds,3); echo "<p>\n<strong>The Human Thioredoxin DNA coding sequence</strong><br>\n<span style=\"font-family:courier;\">$thio_human_cds</span></p>\n"; echo "<p>\n<strong>The translation of the individual codons</strong><br>\n"; foreach($codons as $codon){ $translated_codon = $genetic_code[$codon]; echo "<span style=\"font-family:courier;\">$codon translates to $translated_codon</span><br>\n"; if($translated_codon == "Stop"){break;} $translated_sequence = $translated_sequence.$translated_codon; } echo "</p>\n<p><strong>The Human Thioredoxin protein sequence</strong><br><span style=\"font-family:courier;\">$translated_sequence</span></p>"; ?> |
This is the full output of the code above:
The Human Thioredoxin DNA coding sequence
ATGGTGAAGCAGATCGAGAGCAAGACTGCTTTTCAGGAAGCCTTGGACGCTGCAGGTGATAAACTTGTAGTAGTTGACTTCTCAGCCACGTGGTGTGGGCCTTGCAAAATGATCAAGCCTTTCTTTCATTCCCTCTCTGAAAAGTATTCCAACGTGATATTCCTTGAAGTAGATGTGGATGACTGTCAGGATGTTGCTTCAGAGTGTGAAGTCAAATGCATGCCAACATTCCAGTTTTTTAAGAAGGGACAAAAGGTGGGTGAATTTTCTGGAGCCAATAAGGAAAAGCTTGAAGCCACCATTAATGAATTAGTCTAA
The translation of the individual codons
ATG translates to M
GTG translates to V
AAG translates to K
CAG translates to Q
ATC translates to I
GAG translates to E
AGC translates to S
AAG translates to K
ACT translates to T
GCT translates to A
TTT translates to F
CAG translates to Q
GAA translates to E
GCC translates to A
TTG translates to L
GAC translates to D
GCT translates to A
GCA translates to A
GGT translates to G
GAT translates to D
AAA translates to K
CTT translates to L
GTA translates to V
GTA translates to V
GTT translates to V
GAC translates to D
TTC translates to F
TCA translates to S
GCC translates to A
ACG translates to T
TGG translates to W
TGT translates to C
GGG translates to G
CCT translates to P
TGC translates to C
AAA translates to K
ATG translates to M
ATC translates to I
AAG translates to K
CCT translates to P
TTC translates to F
TTT translates to F
CAT translates to H
TCC translates to S
CTC translates to L
TCT translates to S
GAA translates to E
AAG translates to K
TAT translates to Y
TCC translates to S
AAC translates to N
GTG translates to V
ATA translates to I
TTC translates to F
CTT translates to L
GAA translates to E
GTA translates to V
GAT translates to D
GTG translates to V
GAT translates to D
GAC translates to D
TGT translates to C
CAG translates to Q
GAT translates to D
GTT translates to V
GCT translates to A
TCA translates to S
GAG translates to E
TGT translates to C
GAA translates to E
GTC translates to V
AAA translates to K
TGC translates to C
ATG translates to M
CCA translates to P
ACA translates to T
TTC translates to F
CAG translates to Q
TTT translates to F
TTT translates to F
AAG translates to K
AAG translates to K
GGA translates to G
CAA translates to Q
AAG translates to K
GTG translates to V
GGT translates to G
GAA translates to E
TTT translates to F
TCT translates to S
GGA translates to G
GCC translates to A
AAT translates to N
AAG translates to K
GAA translates to E
AAG translates to K
CTT translates to L
GAA translates to E
GCC translates to A
ACC translates to T
ATT translates to I
AAT translates to N
GAA translates to E
TTA translates to L
GTC translates to V
TAA translates to Stop
The Human Thioredoxin protein sequence
MVKQIESKTAFQEALDAAGDKLVVVDFSATWCGPCKMIKPFFHSLSEKYSNVIFLEVDVDDCQDVASECEVKCMPTFQFFKKGQKVGEFSGANKEKLEATINELV
A few points are worth noting in respect to our DNA sequence to protein sequence translation script above:
- We use a “break” statement, inside the foreach loop that cycles sequentially through each codon, that is executed if we find a “stop” codon in the sequence, as we do not wish to add a “Stop” string in our translated sequence but rather stop the translation job and exit the foreach cycle. Indeed executing a “break” statement within a cycle will stop it and the code that follows the cycle in the script will be executed. This is in contrast with die(), that will entirely terminate the script execution, as we have seen in the PHP conditional statements section earlier in this chapter.
- There are actually some issues with the output as it is now in the script. Specifically, the initial DNA sequence is a very long uninterrupted string and will normally force an horizontal scrolling in the web page in order to be able to see it fully. This is not happening here, in this very page, as the WordPress template used will prevent that. If you however execute the translation script above in a standalone page, you will see that an horizontal scrolling bar will be present, which is not nice. There are of course ways to format the sequence before giving it in output to a webpage, inserting break tags – for example every 80 characters – so as to have a nice display and avoid horizontal scrolling. This was not implemented in this specific example.
- The $translated_sequence variable is declared as empty before the foreach cycle, and then filled up with translation results during the cycle. This is a classical way to proceed: declare an empty string or empty array before the start of a cycle and then fill it up during the cycle. Take note.
Classifying amino-acids in a peptide or protein sequence according to their nature (nonpolar, polar, basic, acidic)
Let us use the ability that we have acquired in this section to split a sequence into individual amino-acids or nucleotides to classify all the amino-acids of a peptide or protein sequence according to their nature, expanding on the example given at the end of the previous section.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 |
<?php $nonpolar ="FLIMVPAWG"; // A string made by all the nonpolar amino-acids in single letter notation $polar = "STYCQN"; // Polar amino-acids $basic = "HKR"; // Basic amino-acids $acidic = "DE"; // Acidic amino-acids $peptide = "MVKQIESKTAFQEALDAAGDKLVVVDF"; // Splitting the peptide in an array of the individual amino-acids $single_aminoacids = str_split($peptide,1); // In the following array $aminoacids_classified we will collect each // aminoacid AND it's nature as a mini sub array of 2 elements // [(aminoacid,nature),(aminoacid,nature),.....] // in order to then provide an output $aminoacids_classified = array(); // Let's start cycling through the amino-acids array foreach($single_aminoacids as $aminoacid){ if(strrchr($nonpolar, $aminoacid)){ $aminoacids_classified[] = array($aminoacid, "nonpolar"); } elseif(strrchr($polar, $aminoacid)){ $aminoacids_classified[] = array($aminoacid, "polar"); } elseif(strrchr($basic, $aminoacid)){ $aminoacids_classified[] = array($aminoacid, "basic"); } elseif(strrchr($acidic, $aminoacid)){ $aminoacids_classified[] = array($aminoacid, "acidic"); } else{ // We leave the possibility open to encounter an unknown character we have not classified $aminoacids_classified[] = array($aminoacid, "unclassified"); } } // We now provide an HTML output echo "<p>\n<strong>Here is a full listing of the amino-acids in our sequence</strong>\n<ol>\n"; foreach($aminoacids_classified as $aa_result){ echo "<li>".$aa_result[0]." => ".$aa_result[1]."</li>\n"; } echo "</ol>\n</p>"; ?> |
This is the output of the script:
Here is a full listing of the amino-acids in our sequence
- M => nonpolar
- V => nonpolar
- K => basic
- Q => polar
- I => nonpolar
- E => acidic
- S => polar
- K => basic
- T => polar
- A => nonpolar
- F => nonpolar
- Q => polar
- E => acidic
- A => nonpolar
- L => nonpolar
- D => acidic
- A => nonpolar
- A => nonpolar
- G => nonpolar
- D => acidic
- K => basic
- L => nonpolar
- V => nonpolar
- V => nonpolar
- V => nonpolar
- D => acidic
- F => nonpolar
Chapter Sections
[pagelist include=”435″]
[siblings]